The Alper Lab rewires cellular metabolism to produce next-generation biochemicals,
polymer precursors, fuels, and pharmaceuticals. In doing so, we are addressing global
challenges including sustainability, human health, circular bio-economies, and clean energy/energy security.
We rewire cells by uniquely merging the areas of synthetic biology, directed evolution, and pathway engineering.
To complement these efforts, we have developed many synthetic genetic toolkits, high-throughput screening techniques, evolution schemes, and culturing platforms.
Our mission is to enable advanced biomanufacturing using rewired host organisms.
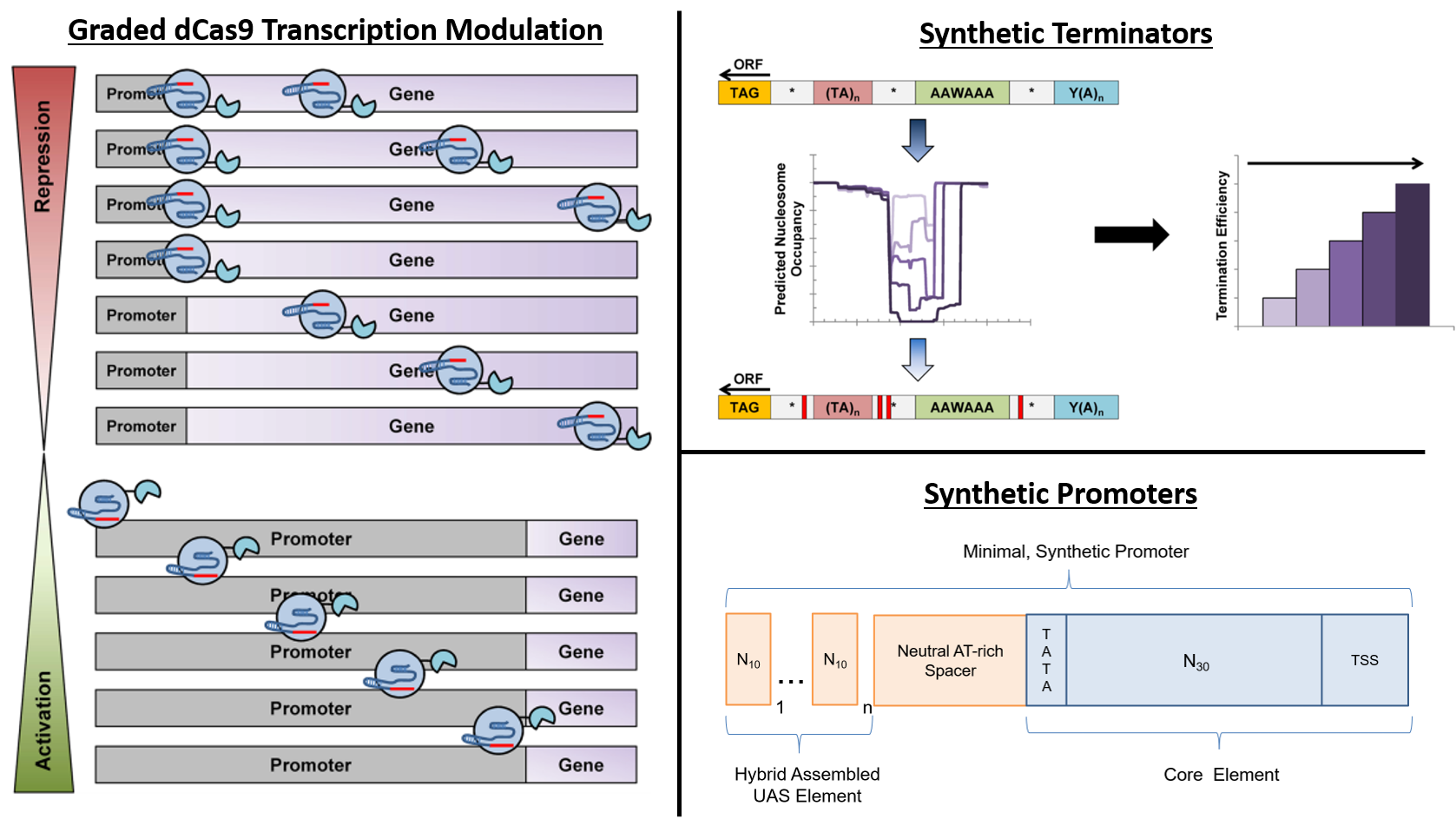
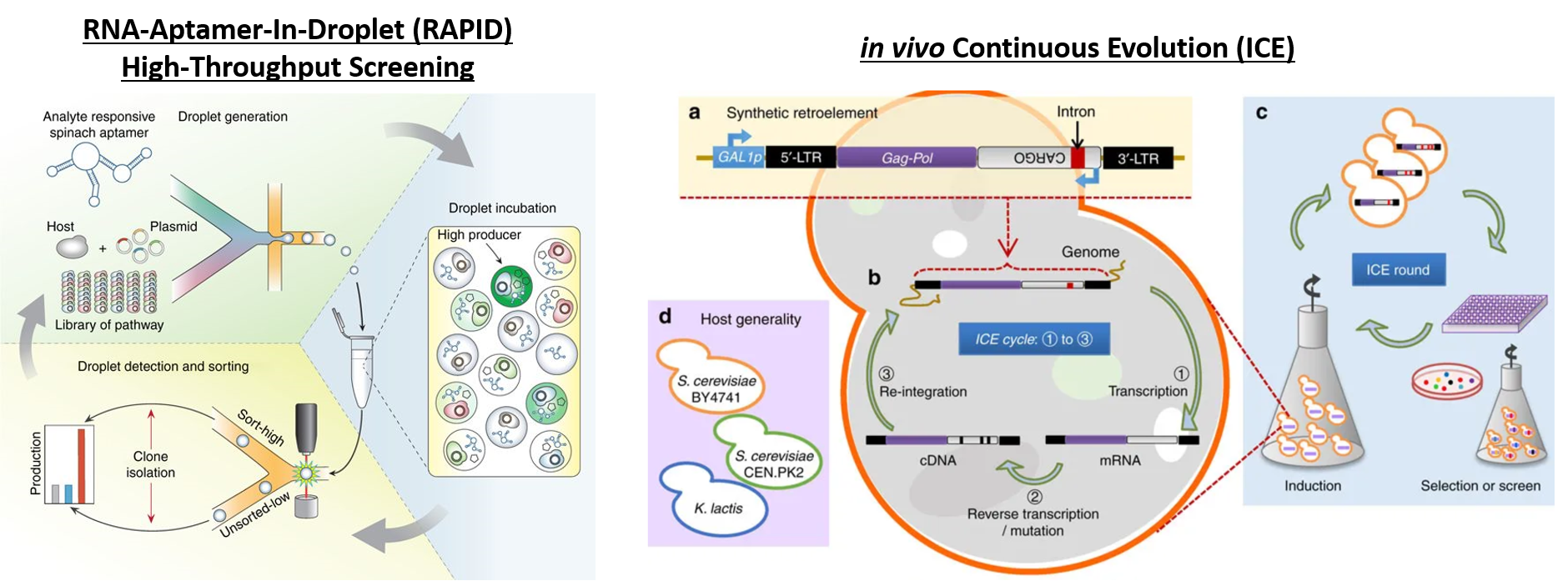
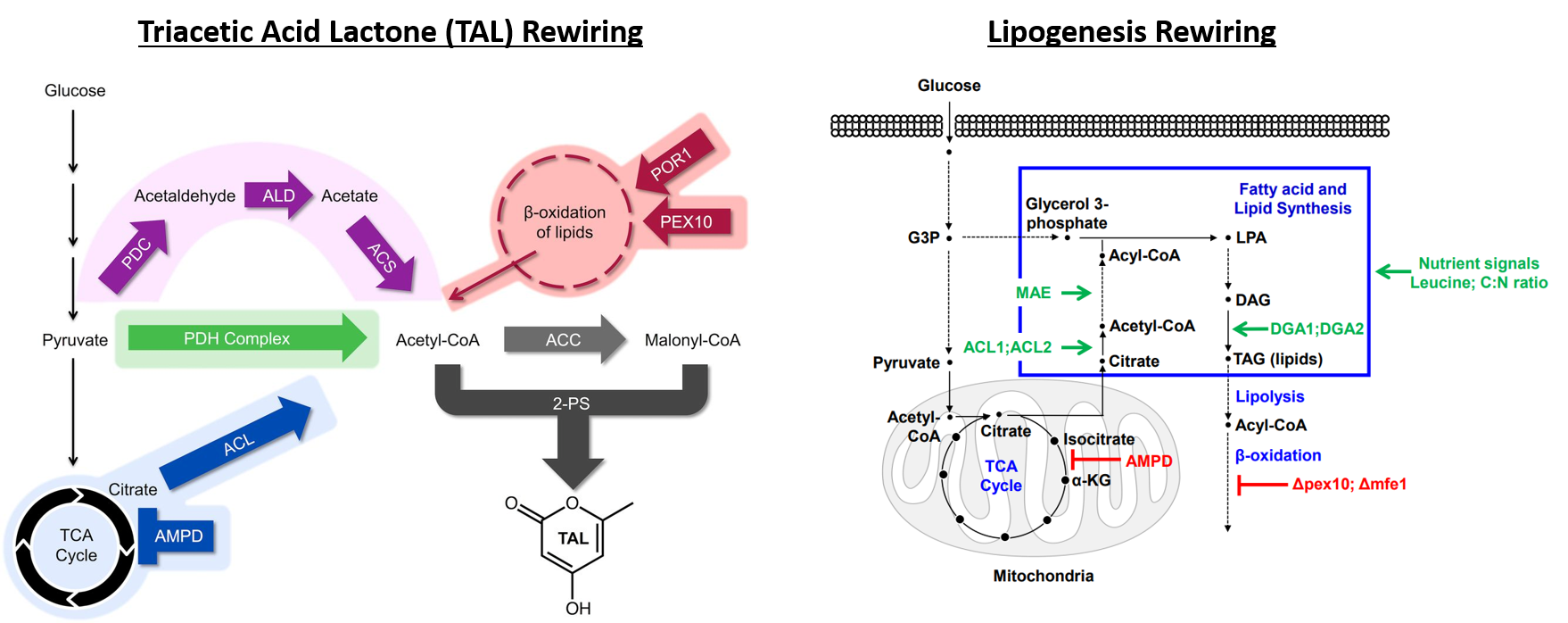
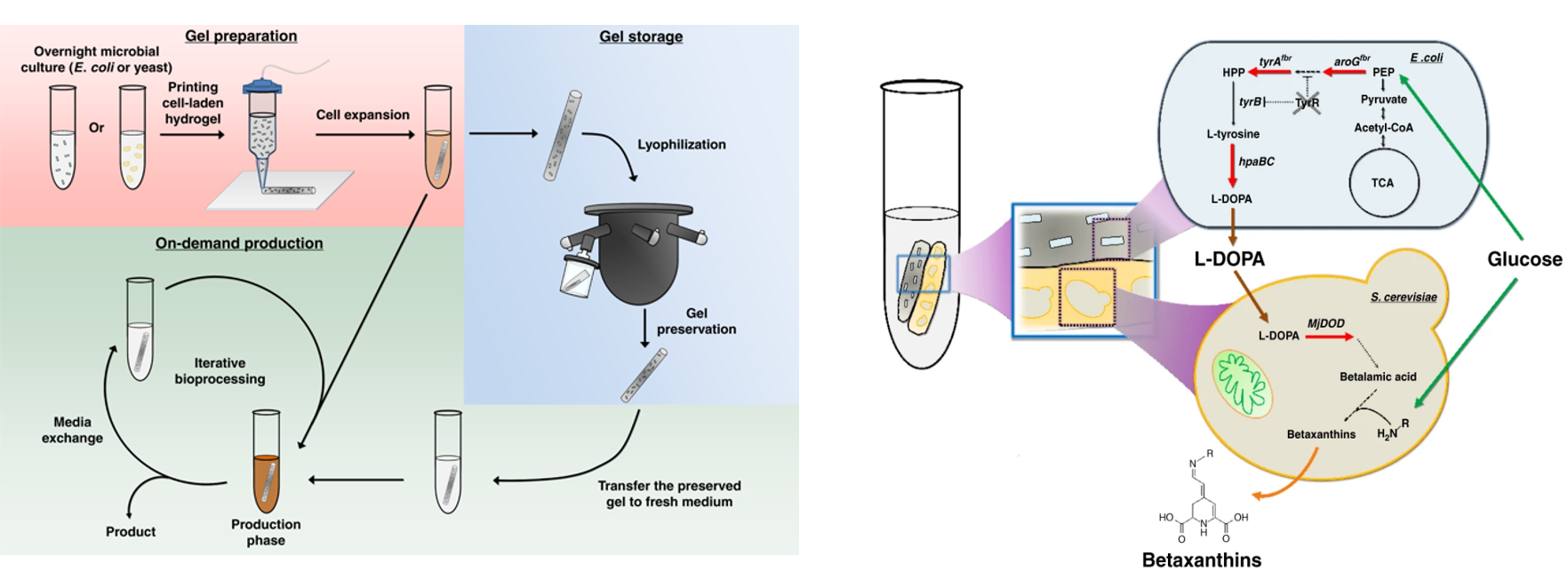
Deaner and Alper, 2017. Systematic Testing of Enzyme Perturbation Sensitivities via Graded dCas9 Modulation. Metabolic Engineering. Link to paper Morse, et al., 2017. Yeast terminator function can be modulated and designed based on predictions of nucleosome occupancy. ACS Synthetic Biology. Link to paper Redden and Alper, 2015. The development and characterization of synthetic minimal yeast promoters. Nature Communications. Link to paper Wagner, et al., 2018. A comparative analysis of single cell and droplet-based FACS for improving production phenotypes: riboflavin overproduction in Yarrowia lipolytica. Metabolic Engineering. Link to paper Abatemarco, et al., 2017. RNA-Aptamer-In-Droplet (RAPID) High-Throughput Screening of Secretory Phenotypes Nature Communications. Link to paper Crook, et al., 2016. in vivo continuous evolution of genes and pathways in yeast. Nature Communications. Link to paper Cordova, et al., 2019. Direct production of fatty alcohols from glucose using engineered strains of Yarrowia lipolytica. Metabolic Engineering Communications. Link to paper Markham, et al., 2019. Rewiring Yarrowia lipolytica toward triacetic acid lactone for novel materials generation. PNAS. Link to paper UT Press Release Blazeck, et al., 2014. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nature Communications. Link to paper 2014 UT Press Release 2015 UT Press Release Johnston, et al., 2020. Compartmentalized microbes and co-cultures in hydrogels for on-demand bioproduction and preservation. Nature Communications. Link to paper UT Press Release